- Publication
- Apr 1, 2024
Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity
Carney, et al. Bioorganic & Medicinal Chemistry, 2024
- Publication
- Mar 12, 2024
Lead optimization of small molecule ENL YEATS inhibitors to enable in vivo studies: Discovery of TDI-11055
Michino M, et al. ACS Med. Chem. Lett., 2024, 15, 4, 524–532
- Publication
- Feb 21, 2024
Structure-guided engineering of immunotherapies targeting TRBC1 and TRBC2 in T cell malignancies
Ferrari, et al. Nat Commun, 2024, 15, 1583
- Publication
- Feb 20, 2024
Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors
Wittlinger, et al. Communications Chemistry, 2024, 7, 38
- Publication
- Feb 19, 2024
Structure–activity relationship of a pyrrole based series of PfPKG inhibitors as anti-malarials
Gilleran JA, et al. Journal of Medicinal Chemistry, 2024, 67(5), 3467–3503
- Publication
- Feb 1, 2024
Enabling Structure-Based Drug Discovery Utilizing Predicted Models
Miller, et al. Cell, 2024, 187(3), 521-525
- Publication
- Jan 19, 2024
Enumerable libraries and accessible chemical space in drug discovery
Knehans T, et al. Computational Drug Discovery: Methods and Applications, 2024, Chapter 14
- Publication
- Dec 15, 2023
Using AlphaFold and Experimental Structures for the Prediction of the Structure and Binding Affinities of GPCR Complexes via Induced Fit Docking and Free Energy Perturbation
Dilek Coskun, et al. J. Chem. Theory Comput., 2023
- Publication
- Dec 5, 2023
Basic Residues at Position 11 of α-Conotoxin LvIA Influence Subtype Selectivity between α3β2 and α3β4 Nicotinic Receptors via an Electrostatic Mechanism
Haufe, et al. ACS Chem. Neurosci, 2023
- Publication
- Dec 1, 2023
Predictive modeling of concentration-dependent viscosity behavior of monoclonal antibody solutions using artificial neural networks
Schmitt, et al. MAbs, 2023, 15(1), 2169440
- Publication
- Nov 28, 2023
Free Energy Perturbation Approach for Accurate Crystalline Aqueous Solubility Predictions
Hong, et al. J. Med. Chem., 2023
- Publication
- Nov 17, 2023
Geometric Deep Learning for Structure-Based Ligand Design
Powers, et al. ACS Central Science, 2023
Webinars
 Webinar
Life Science
Webinar
Life Science
- Apr 23, 2025
Computational strategies for discovering and optimizing RNA- and DNA-targeting molecules
In this webinar, we will showcase how Schrödinger’s advanced computational solutions are enabling accurate and efficient targeting of RNA and DNA systems by small molecules.
 Webinar
Life Science
Webinar
Life Science
- Apr 16, 2025
Schrödinger デジタル創薬セミナー: Into the Clinic~計算化学がもたらす創薬プロセスの変貌~第16回
Accelerating protein degrader discovery: Computational strategies for degrader design and optimization
 Webinar
Life Science
Materials Science
Webinar
Life Science
Materials Science
- Apr 8, 2025
Accelerating pharmaceutical formulations using machine learning approaches
In this webinar, we will demonstrate how Schrödinger’s integrated ML- and physics-based approaches are transforming pharmaceutical formulation design.
Documentation
- Documentation
Online Help and Documentation
An online resource of information and instruction on how to use Schrödinger software including user manuals, panel descriptions, installation guides, reference sheets, tutorials, and more.
 Documentation
Life Science
Documentation
Life Science
Tutorials
- Tutorial
BACE1 Inhibitor Design Using Free Energy Perturbation
Prepare, run, and analyze a free energy perturbation (FEP) simulation for a series of BACE1 inhibitors using FEP+.
- Tutorial
Ligand Binding Pose Prediction for FEP+ using Core-Constrained Docking
Generate starting poses for FEP simulations for a series of BACE1 inhibitors using core constrained docking
- Tutorial
Introduction to MD Simulations with Desmond
Prepare, run, and perform simple analysis on an all-atom MD simulation with Desmond.
Training Videos
 Video
Life Science
Video
Life Science
Getting Going with Maestro BioLuminate
A free video series introducing the basics of using Maestro Bioluminate.
 Video
Life Science
Video
Life Science
 Video
Life Science
Video
Life Science
A Sneak Peek into Renumbering Residues and the Project Table in BioLuminate
The sixth video in the Getting Going with Maestro BioLuminate Video Series: renaming chains and residues, the Project Table.
Publications
- Publication
- Mar 5, 2025
A robust crystal structure prediction method to support small molecule drug development with large scale validation and blind study
Zhou, et al. Nature Communications, 2025, 16, 2210
- Publication
- Feb 28, 2025
Exploiting solvent exposed salt-bridge interactions for the discovery of potent inhibitors of SOS1 using free-energy perturbation simulations
Leffler, et al. ACS Medicinal Chemistry Letters, 2025
- Publication
- Feb 20, 2025
Predicting Resistance to Small Molecule Kinase Inhibitors
Nagarajan, et al. Journal of Chemical Information and Modeling, 2025, Preprint
Case Studies
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
White Papers
 White Paper
Life Science
White Paper
Life Science
- Oct 29, 2024
20 Years of Glide: A Legacy of Docking Innovation and the Next Frontier with Glide WS
Glide has long set the gold standard for commercial molecular docking software due to its robust performance in both binding mode prediction and empirical scoring tasks, ease of use, and tight integration with Schrödinger’s Maestro interface and molecular discovery workflows.
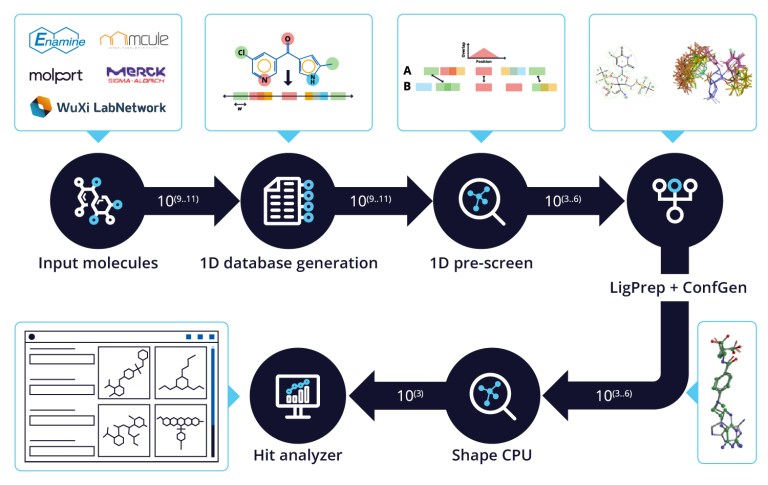 White Paper
Life Science
White Paper
Life Science
 White Paper
Life Science
White Paper
Life Science
Quick Reference Sheets

Latest insights from Extrapolations blog
Training & Resources
Online certification courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Free learning resources
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.