- Publication
- Oct 2, 2024
Predicting the Release Mechanism of Amorphous Solid Dispersions: A Combination of Thermodynamic Modeling and In Silico Molecular Simulation
Walter, et al. Pharmaceutics, 2024, 16(10), 1292
- Publication
- Sep 27, 2024
A method for treating significant conformational changes in alchemical free energy simulations of protein–ligand binding
Liao J, et al. J. Chem. Theory Comput., 2024, 20, 19, 8609–8623
- Publication
- Aug 23, 2024
Novel druggable space in human KRAS G13D discovered using structural bioinformatics and a P-loop targeting monoclonal antibody
Jungholm O et al., Scientific Reports, 2024, 14(1), 19656
- Publication
- Aug 20, 2024
Harnessing free energy calculations to achieve kinome-wide selectivity in drug discovery campaigns: Wee1 case study
Knight J, et al. ChemRxiv, 2024, Preprint
- Publication
- Aug 15, 2024
Robust prediction of relative binding energies for protein–protein complex mutations using free energy perturbation calculations
Sampson, et al. Journal of Molecular Biology, 2024, 436, 16, 168640
- Publication
- Aug 15, 2024
Coarse-grained simulation of mRNA-loaded lipid nanoparticle self-assembly
Grzetic, et al. Molecular Pharmaceutics, 2024, 21, 9, 4747–4753
- Publication
- Aug 13, 2024
OPLS5: Addition of polarizability and improved treatment of metals
Damm, et al. ChemRxiv, 2024
- Publication
- Aug 2, 2024
Quantum chemical package Jaguar: A survey of recent developments and unique features
Cao, et al. The Journal of Chemical Physics, 2024, 161(5), 052502
- Publication
- Jun 24, 2024
Study of key residues in MERS-CoV and SARS-CoV-2 main proteases for resistance against clinically applied inhibitors nirmatrelvir and ensitrelvir
Krismer L et al., Npj Viruses, 2024, 2(1), 23
- Publication
- Jun 19, 2024
Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants
Mittal A, et al. Nature Structural & Molecular Biology, 2024
- Publication
- Apr 23, 2024
Incorporation of multiple β2-hydroxy acids into a protein in vivo using an orthogonal aminoacyl-tRNA synthetase
Hamlish NX, et al. ACS Central Science, 2024, 10(5), 1044–1053
- Publication
- Apr 23, 2024
FEP augmentation as a means to solve data paucity problems for machine learning in chemical biology
Burger PB, et al. J. Chem. Inf. Model., 2024, 64(9), 3812–3825
Webinars
 Webinar
Life Science
Webinar
Life Science
- Apr 23, 2025
Computational strategies for discovering and optimizing RNA- and DNA-targeting molecules
In this webinar, we will showcase how Schrödinger’s advanced computational solutions are enabling accurate and efficient targeting of RNA and DNA systems by small molecules.
 Webinar
Life Science
Webinar
Life Science
- Apr 16, 2025
Schrödinger デジタル創薬セミナー: Into the Clinic~計算化学がもたらす創薬プロセスの変貌~第16回
Accelerating protein degrader discovery: Computational strategies for degrader design and optimization
 Webinar
Life Science
Materials Science
Webinar
Life Science
Materials Science
- Apr 8, 2025
Accelerating pharmaceutical formulations using machine learning approaches
In this webinar, we will demonstrate how Schrödinger’s integrated ML- and physics-based approaches are transforming pharmaceutical formulation design.
Documentation
- Documentation
Online Help and Documentation
An online resource of information and instruction on how to use Schrödinger software including user manuals, panel descriptions, installation guides, reference sheets, tutorials, and more.
 Documentation
Life Science
Documentation
Life Science
Tutorials
- Tutorial
BACE1 Inhibitor Design Using Free Energy Perturbation
Prepare, run, and analyze a free energy perturbation (FEP) simulation for a series of BACE1 inhibitors using FEP+.
- Tutorial
Ligand Binding Pose Prediction for FEP+ using Core-Constrained Docking
Generate starting poses for FEP simulations for a series of BACE1 inhibitors using core constrained docking
- Tutorial
Introduction to MD Simulations with Desmond
Prepare, run, and perform simple analysis on an all-atom MD simulation with Desmond.
Training Videos
 Video
Life Science
Video
Life Science
Getting Going with Maestro BioLuminate
A free video series introducing the basics of using Maestro Bioluminate.
 Video
Life Science
Video
Life Science
 Video
Life Science
Video
Life Science
A Sneak Peek into Renumbering Residues and the Project Table in BioLuminate
The sixth video in the Getting Going with Maestro BioLuminate Video Series: renaming chains and residues, the Project Table.
Publications
- Publication
- Mar 5, 2025
A robust crystal structure prediction method to support small molecule drug development with large scale validation and blind study
Zhou, et al. Nature Communications, 2025, 16, 2210
- Publication
- Feb 28, 2025
Exploiting solvent exposed salt-bridge interactions for the discovery of potent inhibitors of SOS1 using free-energy perturbation simulations
Leffler, et al. ACS Medicinal Chemistry Letters, 2025
- Publication
- Feb 20, 2025
Predicting Resistance to Small Molecule Kinase Inhibitors
Nagarajan, et al. Journal of Chemical Information and Modeling, 2025, Preprint
Case Studies
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
White Papers
 White Paper
Life Science
White Paper
Life Science
- Oct 29, 2024
20 Years of Glide: A Legacy of Docking Innovation and the Next Frontier with Glide WS
Glide has long set the gold standard for commercial molecular docking software due to its robust performance in both binding mode prediction and empirical scoring tasks, ease of use, and tight integration with Schrödinger’s Maestro interface and molecular discovery workflows.
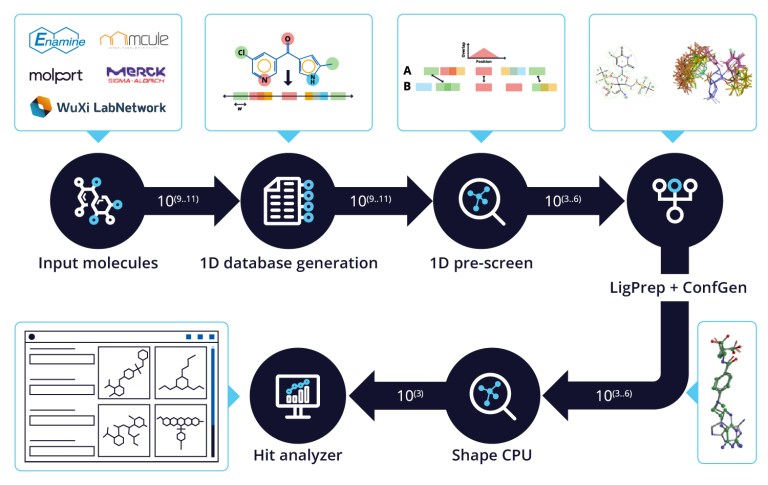 White Paper
Life Science
White Paper
Life Science
 White Paper
Life Science
White Paper
Life Science
Quick Reference Sheets

Latest insights from Extrapolations blog
Training & Resources
Online certification courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Free learning resources
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.