 Webinar
Life Science
Webinar
Life Science
- Aug 31, 2021
Limited Experimental Data? No Problem: Machine Learning and Physics in Preclinical Drug Discovery
In this webinar, we provide three practical examples of the application of machine learning in active drug discovery programs.
 Webinar
Life Science
Webinar
Life Science
 Webinar
Life Science
Webinar
Life Science
- Jul 14, 2021
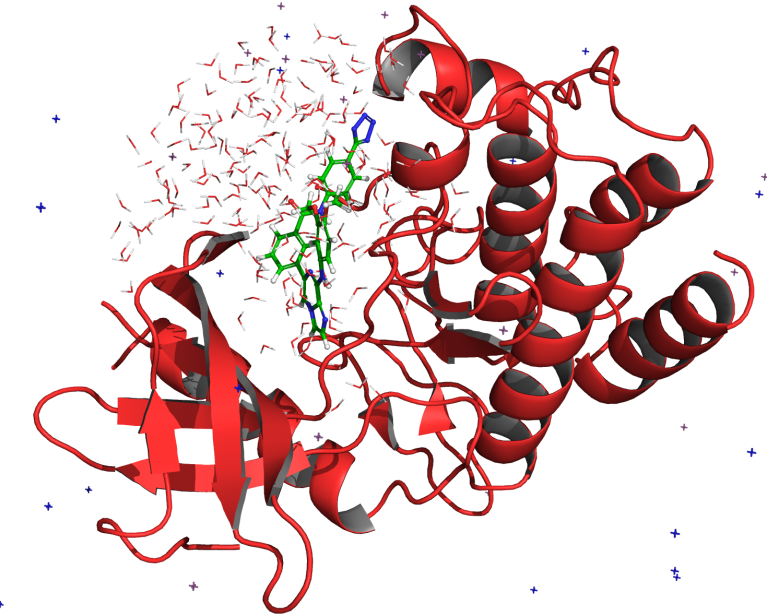
SARS-CoV-2 main protease inhibitors: Physics-based approaches to the discovery of COVID-19 antivirals
In this webinar, we focus on the medicinal and computational chemistry driven discovery of main protease inhibitors.
 Webinar
Life Science
Webinar
Life Science
- Nov 5, 2016
An Introduction to Automating Workflows with KNIME
In this webinar, we give a brief overview on how to get started with KNIME and use Schrödinger Nodes to create workflows for use within KNIME and Maestro.
 Webinar
Life Science
Webinar
Life Science
- Jun 15, 2021
Design and Optimization of Biologics Driven by Physics-Based Computational Modeling
In this webinar, we address the important role that computational modeling can play in accelerating the discovery and development of novel biologics.
 Webinar
Life Science
Webinar
Life Science
 Webinar
Life Science
Webinar
Life Science
- Nov 5, 2018
Creating workflows with KNIME – Beyond the basics
In this webinar, we introduce some more advanced concepts to help you create more complex workflows.
 Webinar
Life Science
Webinar
Life Science
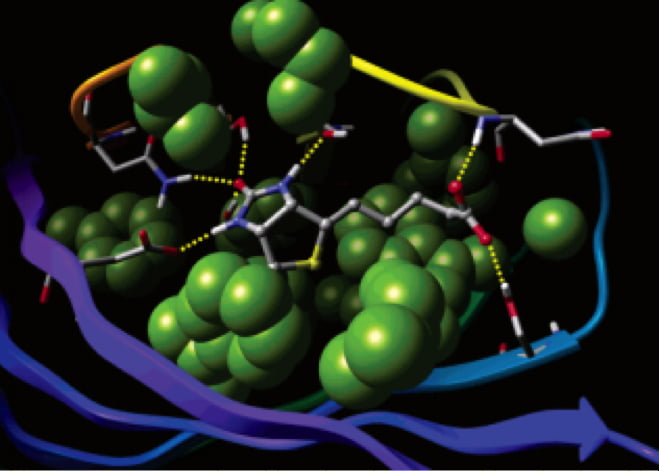
- Feb 23, 2021
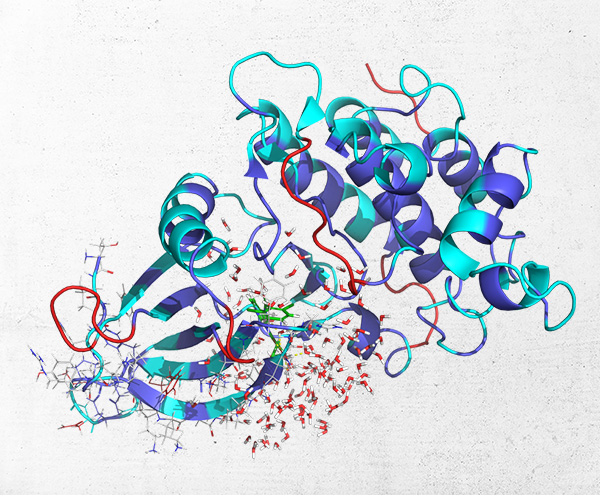
Improving Protein-Ligand Modeling into Cryo-EM Data and the use of those Models in Drug Discovery Efforts
Producing an accurate atomic model of protein-ligand interactions from the data generated by cryo-electron microscopy is often a challenging problem due to a combination of the noise in the experiment and the dynamic nature of protein-ligand binding.
 Webinar
Life Science
Webinar
Life Science
- Jul 16, 2020
Computer-aided Formulation Development for Small-molecule Drugs
In this webinar, we discuss the key areas of formulations science powered by the latest computation chemistry technology, including analysis of chemical processes, assessment of intermolecular interactions in particle/drug systems, and automated in silico property characterization.
 Webinar
Life Science
Webinar
Life Science
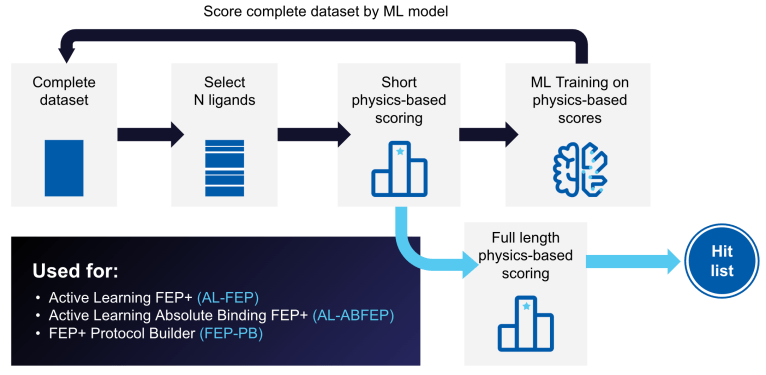
- Jul 13, 2020
Active Learning Glide – Screen Billions of Compounds Efficiently and Cost Effectively
In this webinar, we illustrate how using an Active Learning approach combined with Glide enables cost effective and accurate screening of billion compound libraries.
 Webinar
Life Science
Webinar
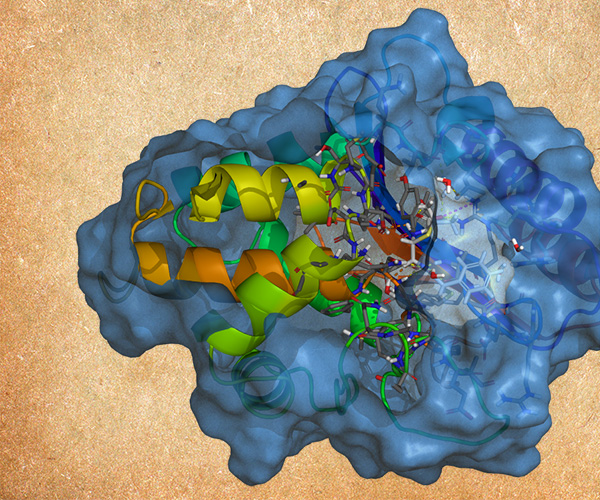
Life Science
- Jul 8, 2020
Enzymes by Design: Structure-based Methods for Modeling Enzymes
An overview of how the Schrödinger technology can be used to optimize enzymes using structure-based rational design.
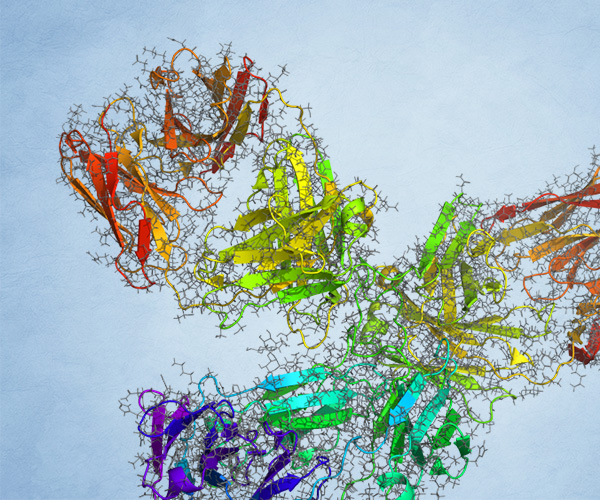 Webinar
Life Science
Webinar
Life Science
- May 27, 2020
Antibody modeling with the Schrödinger Platform
This webinar presents the tools available in BioLuminate to model antibody structures, covering homology modeling, humanization, antigen-antibody docking, liability prediction, and in silico mutations.
Webinars
 Webinar
Life Science
Webinar
Life Science
- Apr 23, 2025
Computational strategies for discovering and optimizing RNA- and DNA-targeting molecules
In this webinar, we will showcase how Schrödinger’s advanced computational solutions are enabling accurate and efficient targeting of RNA and DNA systems by small molecules.
 Webinar
Life Science
Webinar
Life Science
- Apr 16, 2025
Schrödinger デジタル創薬セミナー: Into the Clinic~計算化学がもたらす創薬プロセスの変貌~第16回
Accelerating protein degrader discovery: Computational strategies for degrader design and optimization
 Webinar
Life Science
Materials Science
Webinar
Life Science
Materials Science
- Apr 8, 2025
Accelerating pharmaceutical formulations using machine learning approaches
In this webinar, we will demonstrate how Schrödinger’s integrated ML- and physics-based approaches are transforming pharmaceutical formulation design.
Documentation
- Documentation
Online Help and Documentation
An online resource of information and instruction on how to use Schrödinger software including user manuals, panel descriptions, installation guides, reference sheets, tutorials, and more.
 Documentation
Life Science
Documentation
Life Science
Tutorials
- Tutorial
BACE1 Inhibitor Design Using Free Energy Perturbation
Prepare, run, and analyze a free energy perturbation (FEP) simulation for a series of BACE1 inhibitors using FEP+.
- Tutorial
Ligand Binding Pose Prediction for FEP+ using Core-Constrained Docking
Generate starting poses for FEP simulations for a series of BACE1 inhibitors using core constrained docking
- Tutorial
Introduction to MD Simulations with Desmond
Prepare, run, and perform simple analysis on an all-atom MD simulation with Desmond.
Training Videos
 Video
Life Science
Video
Life Science
Getting Going with Maestro BioLuminate
A free video series introducing the basics of using Maestro Bioluminate.
 Video
Life Science
Video
Life Science
 Video
Life Science
Video
Life Science
A Sneak Peek into Renumbering Residues and the Project Table in BioLuminate
The sixth video in the Getting Going with Maestro BioLuminate Video Series: renaming chains and residues, the Project Table.
Publications
- Publication
- Mar 5, 2025
A robust crystal structure prediction method to support small molecule drug development with large scale validation and blind study
Zhou, et al. Nature Communications, 2025, 16, 2210
- Publication
- Feb 28, 2025
Exploiting solvent exposed salt-bridge interactions for the discovery of potent inhibitors of SOS1 using free-energy perturbation simulations
Leffler, et al. ACS Medicinal Chemistry Letters, 2025
- Publication
- Feb 20, 2025
Predicting Resistance to Small Molecule Kinase Inhibitors
Nagarajan, et al. Journal of Chemical Information and Modeling, 2025, Preprint
Case Studies
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
White Papers
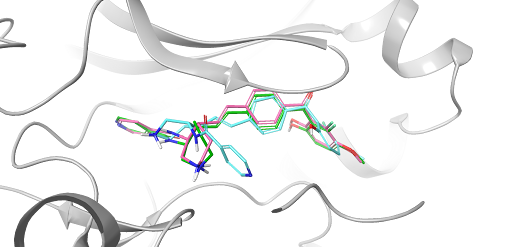 White Paper
Life Science
White Paper
Life Science
- Oct 29, 2024
20 Years of Glide: A Legacy of Docking Innovation and the Next Frontier with Glide WS
Glide has long set the gold standard for commercial molecular docking software due to its robust performance in both binding mode prediction and empirical scoring tasks, ease of use, and tight integration with Schrödinger’s Maestro interface and molecular discovery workflows.
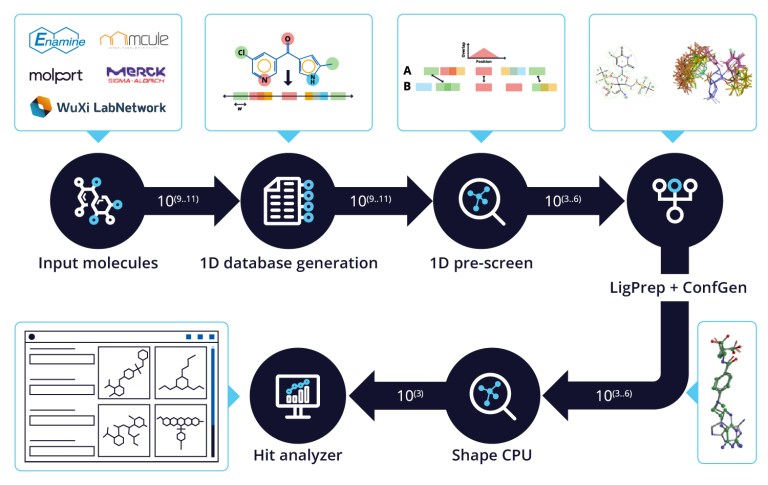 White Paper
Life Science
White Paper
Life Science
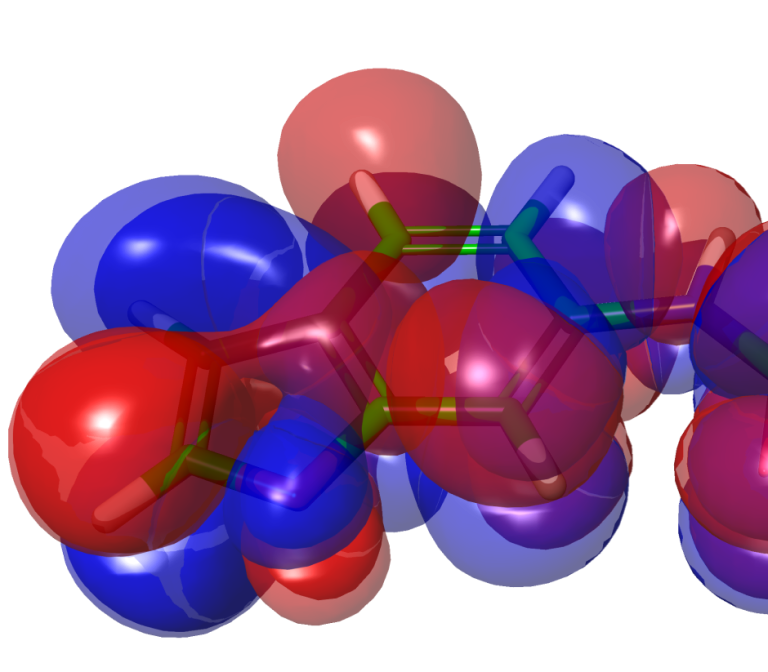 White Paper
Life Science
White Paper
Life Science
Quick Reference Sheets

Latest insights from Extrapolations blog
Training & Resources
Online certification courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Free learning resources
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.