- Publication
- Mar 20, 2001
Solvent Models for Protein-Ligand Binding: Comparison of Implicit Solvent Poisson and Surface Generalized Born Models with Explicit Solvent Simulations
Zhang, et al. J. Comput. Chem., 2001, 22, 591-607
- Publication
- Feb 15, 2001
Large Scale Ab Initio Quantum Chemical Calculations on Biological Systems
Friesner, et al. Accounts of Chemical Research, 2001, 34, 351-358
- Publication
- Jan 1, 2001
Molecular Modeling of Bifunctional Chelate Peptide Conjugates. 1. Copper and Indium Parameters for the AMBER Force Field
Reichert, et al. Inorg. Chem., 2001, 40, 5223-5230
- Publication
- Dec 6, 2000
Hessian-Free Low-Mode Conformational Search for Large-Scale Protein Loop Optimization: Application to c-jun N-Terminal Kinase JNK3
Kolossvary, et al. J. Comp. Chem., 2001, 22, 21-30
- Publication
- Dec 1, 2000
Design, Synthesis and Binding Properties of Novel and Selective 5-HT3 and 5-HT4 Receptor Ligands
Modica, et al. Eur. J. Med. Chem., 2000, 35, Erratum: ibid 2001, 36 287-301
- Publication
- Nov 14, 2000
A Mixed Quantum Mechanics/Molecular Mechanics (QM/MM) Method for Large-scale Modeling of Chemistry in Protein Environments
Murphy, et al. J. Comp. Chem., 2000, 21, 1442-1457
- Publication
- Nov 5, 2000
Protonation states of the chromophore of denatured green fluorescent proteins predicted by ab initio calculations
El Yazal, et al. J. Am. Chem. Soc., 2000, 122, 11411-11415
- Publication
- Oct 24, 2000
Design, Synthesis, and Solution Structure of a Pyrrolinone-Based ‘-Turn Peptidomimetic
Smith, et al. J. Am. Chem. Soc., 2000, 122, 11037-11038
- Publication
- Jun 6, 2000
Structures of Protonated Arginine Dimer and Bradykinin Investigated by Density Functional Theory: Further Support for Stable Gas-Phase Salt Bridges
Strittmatter, et al. J. Phys. Chem. A., 2000, 104, 6069-6076
- Publication
- Jun 5, 2000
Prediction of Drug Solubility from Monte Carlo Simulations
Jorgensen, et al. Bioorg. Med. Chem. Lett., 2000, 10, 1155-8
- Publication
- May 4, 2000
Improved Treatment of Cyclic Beta-Amino Acids and Successful Prediction of Beta-Peptide Secondary Structure using a Modified Force Field: AMBER*C
Christianson, et al. J. Comput. Chem., 2000
- Publication
- Mar 10, 2000
Prediction of Properties from Simulations: Free Energies of Solvation in Hexadecane, Octanol, and Water
Duffy, et al. J. Am. Chem. Soc., 2000, 122, 2878-88
Webinars
 Webinar
Life Science
Webinar
Life Science
- Apr 23, 2025
Computational strategies for discovering and optimizing RNA- and DNA-targeting molecules
In this webinar, we will showcase how Schrödinger’s advanced computational solutions are enabling accurate and efficient targeting of RNA and DNA systems by small molecules.
 Webinar
Life Science
Webinar
Life Science
- Apr 16, 2025
Schrödinger デジタル創薬セミナー: Into the Clinic~計算化学がもたらす創薬プロセスの変貌~第16回
Accelerating protein degrader discovery: Computational strategies for degrader design and optimization
 Webinar
Life Science
Materials Science
Webinar
Life Science
Materials Science
- Apr 8, 2025
Accelerating pharmaceutical formulations using machine learning approaches
In this webinar, we will demonstrate how Schrödinger’s integrated ML- and physics-based approaches are transforming pharmaceutical formulation design.
Documentation
- Documentation
Online Help and Documentation
An online resource of information and instruction on how to use Schrödinger software including user manuals, panel descriptions, installation guides, reference sheets, tutorials, and more.
 Documentation
Life Science
Documentation
Life Science
Tutorials
- Tutorial
BACE1 Inhibitor Design Using Free Energy Perturbation
Prepare, run, and analyze a free energy perturbation (FEP) simulation for a series of BACE1 inhibitors using FEP+.
- Tutorial
Ligand Binding Pose Prediction for FEP+ using Core-Constrained Docking
Generate starting poses for FEP simulations for a series of BACE1 inhibitors using core constrained docking
- Tutorial
Introduction to MD Simulations with Desmond
Prepare, run, and perform simple analysis on an all-atom MD simulation with Desmond.
Training Videos
 Video
Life Science
Video
Life Science
Getting Going with Maestro BioLuminate
A free video series introducing the basics of using Maestro Bioluminate.
 Video
Life Science
Video
Life Science
 Video
Life Science
Video
Life Science
A Sneak Peek into Renumbering Residues and the Project Table in BioLuminate
The sixth video in the Getting Going with Maestro BioLuminate Video Series: renaming chains and residues, the Project Table.
Publications
- Publication
- Mar 5, 2025
A robust crystal structure prediction method to support small molecule drug development with large scale validation and blind study
Zhou, et al. Nature Communications, 2025, 16, 2210
- Publication
- Feb 28, 2025
Exploiting solvent exposed salt-bridge interactions for the discovery of potent inhibitors of SOS1 using free-energy perturbation simulations
Leffler, et al. ACS Medicinal Chemistry Letters, 2025
- Publication
- Feb 20, 2025
Predicting Resistance to Small Molecule Kinase Inhibitors
Nagarajan, et al. Journal of Chemical Information and Modeling, 2025, Preprint
Case Studies
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
 Case Study
Life Science
Case Study
Life Science
White Papers
 White Paper
Life Science
White Paper
Life Science
- Oct 29, 2024
20 Years of Glide: A Legacy of Docking Innovation and the Next Frontier with Glide WS
Glide has long set the gold standard for commercial molecular docking software due to its robust performance in both binding mode prediction and empirical scoring tasks, ease of use, and tight integration with Schrödinger’s Maestro interface and molecular discovery workflows.
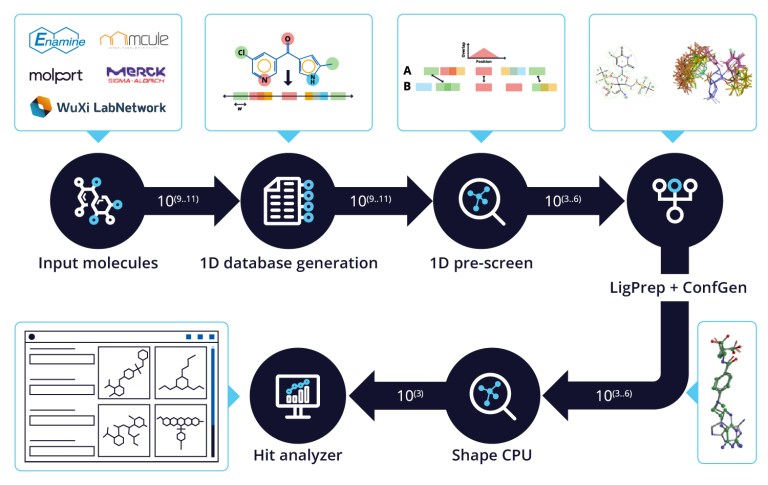 White Paper
Life Science
White Paper
Life Science
 White Paper
Life Science
White Paper
Life Science
Quick Reference Sheets

Latest insights from Extrapolations blog
Training & Resources
Online certification courses
Level up your skill set with hands-on, online molecular modeling courses. These self-paced courses cover a range of scientific topics and include access to Schrödinger software and support.
Free learning resources
Learn how to deploy the technology and best practices of Schrödinger software for your project success. Find training resources, tutorials, quick start guides, videos, and more.